License : Creative Commons Attribution 4.0 International (CC BY-NC-SA 4.0)
Copyright :
Hervé Frezza-Buet,
CentraleSupelec
Last modified : February 15, 2024 11:13
Link to the source : index.md
Table of contents
Risks lecture materials
The theoretical curves corresponding to consistent ERM are this:
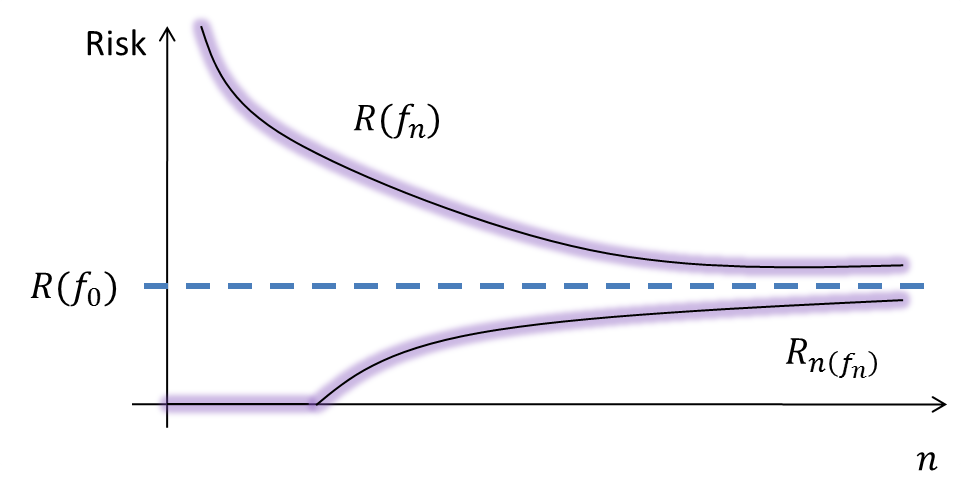
This can be experimented, for example with SVMs.
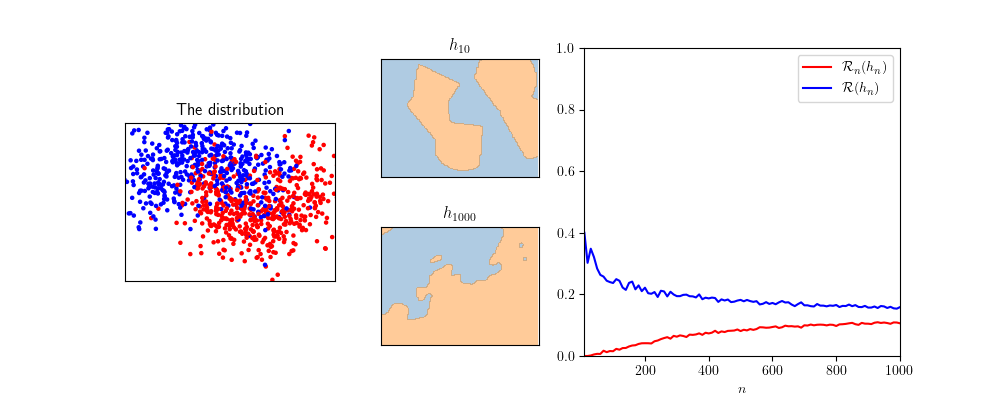
The python source code for such experiments is given below:
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.gridspec as gspec
import sklearn.datasets
import sklearn.svm
import sklearn.tree
# We setup the display
matplotlib.rcParams['text.usetex'] = True
xlim = [-1.5, 2.5]
ylim = [-1.5, 1.5]
fig = plt.figure(figsize=(10,4))
gs = gspec.GridSpec(2, 3,
width_ratios =[4, 3, 6],
height_ratios =[1, 1])
# This function plots the dataset and the decision boundary.
def plot_classifier(classifier, X, y, gridspec, title, draw_contour, draw_samples, Nsamples=100):
ax = plt.subplot(gridspec)
ax.set_xlim(xlim)
ax.set_ylim(ylim)
ax.set_aspect('equal')
ax.set_title(title)
ax.set_xticks([])
ax.set_yticks([])
if draw_contour:
dx = (xlim[1] - xlim[0])/float(Nsamples)
dy = (ylim[1] - ylim[0])/float(Nsamples)
xx, yy = np.meshgrid(np.arange(xlim[0], xlim[1], dx),
np.arange(ylim[0], ylim[1], dy))
Z = classifier.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
colors=np.array(['#377eb8','#ff7f00'])
plt.contourf(xx, yy, Z, alpha=0.4,
levels=[0, 0.5, 1.0], colors=colors, zorder=1)
if draw_samples:
colors=np.array(['blue','red'])
plt.scatter(X[:,0], X[:,1], color=colors[y], s=5, zorder=2)
# Ok, here we are ready to do machine learing
# R_n is the measure of the amount of errors made by a classifier on a
# dataset.
def empirical_risk(classifier, X, y) :
ypred = classifier.predict(X)
return sum(y != ypred)/float(y.shape[0])
# With artificial distributions (sampler is one such), it is easy to
# generate a new dataset (i.i.d) and to measure the performance of a
# classifier on this dataset. This estimates the real risk (law of
# large numbers), no need for cross-validation here.
def real_risk(classifier, sampler, N=1000) :
X, y = sampler(N)
return empirical_risk(classifier, X, y)
# This is the main.
oracle_noise = 0.4 # Set oracle noise (classes get mixed with noise)
Nstep = 10 # Plot curves every Nstep dataset size.
Nmin = Nstep # Min dataset size
Nmax = 1000 # Max dataset size
nb_average = 20 # For each n, risks are averaged over nb_average runs.
sampler = lambda n: sklearn.datasets.make_moons(n, noise=oracle_noise)
experiment = ['SVM', 'Tree'][2]
if experiment == 'SVM':
sigma = .15 # .05 overfits, 2 has an inductive bias.
classifier = sklearn.svm.SVC(C=10,
kernel='rbf', gamma=.5/(sigma*sigma),
tol=1e-5,
decision_function_shape='ovo')
if experiment == 'Tree':
max_depth = 100 # Try the parameters, overfitting is not obvious even for deep trees.
classifier = sklearn.tree.DecisionTreeClassifier(max_depth = max_depth,
min_samples_split = 2)
# Let us plot the learner capabilities
X, y = sampler(Nmax)
plot_classifier(None, X, y, gs[:, 0], 'The distribution', False, True)
X, y = sampler(Nmin)
classifier.fit(X, y)
plot_classifier(classifier, X, y, gs[0, 1], '$h_{{{}}}$'.format(Nmin), True, False)
X, y = sampler(Nmax)
classifier.fit(X, y)
plot_classifier(classifier, X, y, gs[1, 1], '$h_{{{}}}$'.format(Nmax), True, False)
# Let us compute and plot statistics
risks = []
Ns = [N for N in range(Nmin, Nmax+1, Nstep)]
for N in Ns :
print('Computing {} risks for N = {}'.format(nb_average, N))
stats = []
for k in range(nb_average) :
X, y = sampler(N)
classifier.fit(X, y)
stats.append([empirical_risk(classifier, X, y), real_risk(classifier, sampler, N)])
risks.append(np.average(np.array(stats), axis=0))
risks = np.array(risks)
ax = plt.subplot(gs[:,2])
ax.set_xlim([Nmin, Nmax])
ax.set_ylim([0, 1])
ax.set_xlabel('$n$')
plt.plot(Ns, risks[:,0], 'r-', label='${{{\\cal R}}}_n(h_n)$')
plt.plot(Ns, risks[:,1], 'b-', label='${{{\\cal R}}}(h_n)$')
plt.legend();
# Show all
plt.savefig('risks.png', layout="tight")
print()
print()
print('risks.png saved')
plt.show()
Hervé Frezza-Buet,