License : Creative Commons Attribution 4.0 International (CC BY-NC-SA 4.0)
Copyright :
Jérémy Fix,
CentraleSupelec
Last modified : February 15, 2024 11:13
Link to the source : index.md
Dimensionality reduction
In this practical, we will be experimenting with dimensionality reduction methods (either variable selection or variable extraction) in various contexts :
- experimenting filters and wrappers to reduce the dimensionality of the input data in a classification or regression problem, for hopefully getting faster to compute predictors able to better generalize,
- reducing the dimensionality of some data for visualization, in particular for inspecting how the features of different classes get separated along the depth of a neural network
For doing the labwork
Dimensionality reduction for learning better predictors
In this section, we will be working with dataset whose feature space dimension is really large compared to the number of samples we possess. We aim at experimenting with various dimensionality reduction techniques to assess if these are beneficial with respect to the quality of the predictors we obtain. But “better predictors” is sometimes more than that, “better” might also be understood as sparser leading to faster to evaluate predictors and sometimes easier to interpret ones (for example \(1.000.000\) dimensions boiled down to \(2.000\) or so).
Template code
I provide you with a template code to fill in : featsel_template.py. You should also download and place in the same directory the following two files : stanford_sentiment.py and utils.py. stanford_sentiment.py is an helper for getting the dataset (more about that in the next paragraph) and utils.py provides utilitary functions to cache python objects on the drive (you do not have to use these functions, though they may be helpfull for speeding up some processings).
For easily creating a virtual environment with required dependencies, you can either use the requirements.txt or the conda environment.yml file. For using the requirements file :
python3 -m venv venv
source venv/bin/activate
python -m pip install -r requirements.txt
Dataset
We will experiment with the Stanford 50K reviews dataset (don’t download it by yourself). The dataset contains movie reviews: \(25.000\) in the training set and \(25.000\) in the test set.These reviews are labeled either positive or negative. There is also an additional \(50.000\) unlabeled reviews. We will not use the unlabeled data in this practical though some techniques could make use of them (e.g. TfIdf). Below is an example of positive review
Is it worth seeing? If you can look past the technical and budgetary limitations, and get into the story, I think you will enjoy this, especially if you’ve actually read the original H G Wells novel. If, however, you are easily put off by cheap production values, you’d best pass on this (unless you’re a MST3K fan). Be warned, however that the film runs a full 3 hours, so I don’t recommend watching it all in one sitting.<br /><br /> BTW: An entirely different version of War of the Worlds (aka “INVASION”) came out on DVD the same month that Spielberg’s hit the theatres: http://imdb.com/title/tt0449040/. This was also made on a budget, but is updated to the present day like the Spielberg film - but it’s much better! And to top it off, Jeff Wayne is making an animated film of his best-selling album from 1978, but that won’t be out until 2007.
extract of pos/2373_7.txt
And now a negative review
Yaaaaaaaaaaaaaawwwwwwwwwwwwwwwwwnnnnnnnnnnnnn! :=8O<br /><br />ZZZZZZZZzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz……….. <=8.<br /><br />Oh, um excuse me, sorry, fell asleep there for a mooment. Now where was I? Oh yes, “The Projected Man”, yes… ZZZZZZZZzzzzzzzzzzzzzzzzzzzzzzz……….. <=8.<br /><br />Ooops, sorry. Yes, “The Projected Man”. Well, it’s a British sci-fi yawnfest about nothing. Some orange-headed guy projects himself on a laser, gets the touch of death. At last he vanishes, the end. Actually, the film’s not even that interesting. Dull, droning, starchy, stiff, and back-breakingly boring, “The Projected Man” is 77 solid minutes of nothing, starring nobody. Dull as dishwater. Dull as doorknob dust. Dull as Ethan Hawke - we’re talking really DULL here, people! But wait, in respect to our dull cousins from across the puddle, the MooCow will now do a proper review for “The Projected Man”:<br /><br />ZZZZZZZZZZzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz………….. <=8.
neg/9740_1.txt
As you may see, texts are not all written by Shakespeare and there is a lot of fancy variabilities. The task is to predict the polarity of the review, a so-called sentiment analysis task.
To access the data, I wrote a script which downloads and loads the text. Take stanford_sentiment.py and you can access the data with this python snippet
traindata = stanford_sentiment.fetch_imdb(subset='train')
unsupdata = stanford_sentiment.fetch_imdb(subset='unsup')
testdata = stanford_sentiment.fetch_imdb(subset='test')
This will download and perform some structural change on the data to speed up its access. However, it may take \(15\) minutes or so. Therefore, I suggest you to download imdb_py3.pkz and to place it into your scikit dataset directory which is by default on linux ~/scikit_learn_data/. Then, interpret stanford_sentiment.py, it should get executed in less than a minute.
Preprocessing
Our data contain a lot of noise with respect to the sentiment analysis task and we should always do our maximum to get rid of it (otherwise you waste time and data to do that cleaning).
Here is a list of useless variability :
- uppercase/lowercase (see the python documentation)
- we can get rid of any URLS of the form http://... and https://...
- we can remove any HTML tags such as < br/>
- we can remove any newline (but that’s not a big deal to keep them)
- we can remove any dates (such as 1978 in the positive review above)
- we can remove any words that contain digits (such as 70s, 19th)
- …
To do that preprocessing, regular expressions are of a great help and I suggest you to use the regular expression python module. The preprocessing function of a single text is to be defined in the def preprocess(txt) function within the template code.
I give you below some options you may consider :
import re
import string
txt = """See here for details http://www.dvdbeaver.com/film/DVDCompare2/kingofmasks.htm.<br /> Anyway <br />Wonderful performances by the two main actors (The King and Doggie) !
This was worth seeing in the 60s. So baaaaaad.
"""
# To lower case
print("To lower case")
print(txt.lower())
# Remove links
print("Removing the links")
url_regexp="https?://[\w\/\.]+"
print(re.sub(url_regexp, ' ', txt))
# Remove the HTML tags <.../>
print("Removing the HTML tags")
print(re.sub('<[ a-zA-Z]*/?>', ' ', txt))
# Remove punctuation
print("Removing the punctuation")
print(re.sub('[%s]' % re.escape(string.punctuation), ' ', txt))
# Remove linebreaks
print("Removing the linebreaks")
print(re.sub('\n', ' ', txt))
# Remove words containing numbers
print("Removing words containing numbers")
print(re.sub('\w*\d\w*', ' ', txt))
# Remove duplicated characters that are present more than 2 times
# like : reaaaaaaally => really
print("Removing duplicated letters")
print(re.sub(r'(.)\1{2,}', r'\1', txt))
amazing, amazingly would be both transformed into amaz by the nltk snowball stemmer.
from sklearn.feature_extraction import text
scikit_stop_words = text.ENGLISH_STOP_WORDS
print(sorted(scikit_stop_words))
# ['a', 'about', 'above', 'across', 'after', 'afterwards', 'again', 'against', 'all', 'almost', 'alone', 'along', 'already', ...
import nltk
import nltk.corpus as corpus
nltk.download('stopwords')
nltk_stop_words = corpus.stopwords.words('english')
print(sorted(nltk_stop_words))
# ['a', 'about', 'above', 'after', 'again', 'against', 'ain', 'all', 'am', 'an', 'and', 'any', 'are', 'aren', "aren't", 'as', 'at', 'be', 'because', 'been', ...
However, be carefull and ensure that your preprocessing does not impede you from predicting the label; for example, both stopwords list contain the word not and, clearly, removing not from the sentence That movie is not good is not a good option.
Once your preprocess function is written, you can call the function below. You can specify the number of concurrent num_jobs to be executed. Set it to \(-1\) to use all your CPUs.
# Load the datasets and preprocess it
traindata, unsup, testdata = preprocess_imdb(num_jobs=num_jobs)
This will apply your preprocessing function on the whole dataset, store the preprocessed data in a cache file named imdb.pkz and return the result. Important if you change the preprocess function, you have to manually remove the imdb.pkz file from the directory where you executed the script in order to preprocess the whole dataset again, otherwise, your program will use the preprocessed data cached on your drive.
A simple baseline
Let us consider a baseline predictor which will be a bag-of-words with n-grams followed by a linear classifier. You must fill-in the definition of simple_model in the template code. Using scikit-learn, implementing this classifier is just a question of sticking together already implemented algorithms.
The bag-of-words with n-grams is implemented as the CountVectorizer. I suggest you to check the example code on the documentation, to let all the parameters at their defaults except for the ngram_range and min_df. For computing unigram, bi-gram, tri-gram and so, you need to set ngram_range to \((1,1)\), \((1,2)\) and \((1,3)\) respectively. The min_df, which is either a float or an int, excludes the words that are not present at least that frequency (for float) or at least that number of time (for int). The min_df can significanlty reduce your vocabulary size. Implement the CountVectorizer and vectorize your data. Do not hesitate to print out vectorized data, and the vocabulary can be obtained from the CountVectorizer.get_feature_names method.
As a note, if you print out one of the vectorized data, you will notice that it is a sparse structure scipy.sparse.csr.csr_matrix which is memory efficient for bag-of-words representation of text documents. Indeed, your documents contain \(50\) words or so with a vocabulary size of \(40.000\) for uni-grams, \(400.000\) for {1-2}-grams and \(800.000\) for {1-3}-grams or more if you do not exclude words that are present less than 2 times. Therefore one document has a very sparse representation.
Once both your training and testing data are vectorized with the bag-of-words, we can introduce the classifier. I suggest to use a simple linear classifier, a LogisticRegression. I suggest you to let the default values for the parameters. This classifier optimizes :
\[ \begin{align} \hat{y_i} &= \frac{1}{1 + \exp(w^T x_i)}\\ \mathcal{L}(w) &= \frac{1}{2} \|w\|_2^2- C\frac{1}{N} \sum_{i=0}^{N-1} (y_i \log(\hat{y_i}) + (1-y_i)\log(1-\hat{y_i}) ) \end{align} \]
\(x_i \in \mathbb{R}^n, y_i \in \{0, 1\}\) being your training data. The hyperparameter \(C\) is the inverse of the regularization coefficient. You could try to find it by GridSearch, given the risk estimated by cross validation but that’s not the point in this practical, we just want a baseline, so keep the defaults. However, I suggest you to put a scaler before the logistic regression, for example a MaxAbsScaler. Finally, I also advise you to set up a scikit Pipeline made of the vectorizer, scaler and the logistic regression. With scikit, you can fit the pipeline, evaluate the metrics (for example the accuracy) and estimate the real risk by cross-validation easily, see test_iris.py for an example and transpose it to your case.
With a {1-2}-gram bag-of-words, you should get a cross validated accuracy of around 90% .
A little question, why do you think it is necessary to put the count vectorizer in the pipeline that is cross-validated ?
Univariate filters
For this section, you should fill in the function imdb_filter in the template code.
Using {1-2}-grams, excluding words present less than \(2\) times, your feature space should be around \(400.000\) dimensions or so and remember we have only \(25.000\) data to train on. We want to use filters and in particular, we will be testing univariate filters to select few of these dimensions. On the input side, we have words that are either present or absent and when are present can be present more than one time. On the other size, we have a binary evaluation, either positive or negative. We can then use either a statistical test or an information measure to test for the independence between the presence/absence of a n-gram and the target. As input, instead of a binary variable presence/absence, you could also consider the count (an integer) of the n-gram in the document.
The process is the following :
- compute for each documnent, the number of occurence of each nègrams
- select the features (n-grams) the most informative about the target
- fit a LogisticRegression with a MaxAbsScaler
The first step is to determine the presence or absence (or count) of n-grams. Here you can still use the CountVectorizer (check the argument binary). The result of fitting and transforming your data with the CountVectorizer are (sparse) arrays whose values are either \(0\) or or \(n \in \mathbb{N}*\)). I suggest you to use the same settings as for the simple model, using {1-2}-grams that are at least present \(2\) times in the dataset. Then, we apply an univariate filter to drop the n-grams that are most independent from the target. I suggest you to try a statistical test such as the \(\chi^2\). An example code adapted from the documentation shows how to select \(k\) features with a \(\chi^2\) test and to know which dimensions are kept:
from sklearn.datasets import load_digits
from sklearn.feature_selection import SelectKBest, chi2
X, y = load_digits(return_X_y=True)
print(X.shape)
# (1797, 64)
selector = SelectKBest(chi2, k=20)
X_new = selector.fit_transform(X, y)
print(X_new.shape)
# (1797, 20)
support_mask = selector.get_support()
print(support_mask)
# [False False False False False True True ...
The support mask is a boolean numpy array which can be used to keep only the selected dimensions within a numpy array. As an example, the snippet below shows how to extract the part of the vocabulary that has been selected by a filter:
counter = CountVectorizer(...)
counter.fit(...)
selector = SelectKBest(..)
selector.fit(...)
selected_dims = selector.get_support() # boolean array
# Extract the words (n-grams) kept by the univariate filter
selected_terms = np.array(counter.get_feature_names())[selected_dims]
Now you are ready to setup a scikit pipeline involving all the required steps for testing the filter approach. Compute the metrics (train/test accuracies), estimate the real risk by cross-validation (remember that test_iris.py illustrates how to do so) and experiment with different number of features for the filter and different regularization coefficient. Also, we can introduce a measure of sparsity of your model as the fraction of dimensions you kept from the original vocabulary and which can be computed with the code we provide by calling the sparsity_scorer function.
The best I was able to reach is a cross-validated risk of \(89\%\) (mean accuracy) with a sparsity of \(83\%\).
Embedded
An embedded algorithm relies on a machine learning algorithm which, by design selects a subset of the features. To name a few methods belonging to this category: decision trees whose depth limits the number of features involved in the decision, support vector machines with an L1 penalty or LASSO whose L1 regularization leads to sparse predictors.
In particular, I propose to use again a LogisticRegression but this time, with a L1 penalty on the weights:
\[ \begin{align} \hat{y_i} &= \frac{1}{1 + \exp(w^T x_i)}\\ \mathcal{L}(w) &= \|w\|_1 - C\frac{1}{N} \sum_{i=0}^{N-1} (y_i \log(\hat{y_i}) + (1-y_i)\log(1-\hat{y_i}) ) \end{align} \]
Again, I advise you to set up a pipeline using a LogisticRegression on a bag-of-word representation, not forgetting the MaxAbsScaler. With this approach, you do not directly specify a number of features to keep. It is a result of the optimization and depends on the regularization coefficient \(1/C\). For small \(C\), the L1 penalty is more important than the classification loss and the optimizer will simply drop all the components. For large \(C\), the L1 penalty is not dominating the total loss and all the dimensions will probably be involved to some extent.
Once your model is fitted on the training set, evaluate the accuracy on the training set and test set, save the vocabulary that has been selected and evaluate the sparsity of your model. For evaluating the sparsity of your model, you can use the sparsity_scorer function that is given to you in the template code. It can directly take in a fitted classifier or pipeline and basically looks for the non null coefficients of your LogisticRegression. Actually, that function also gives you a hint on how to extract the selected vocabulary out of the full vocabulary of your vectorizer.
Finally, run a cross validation (remember that test_iris.py illustrates how to do so) of your algorithm.
Wrappers
The code for this section is expected to be filled in the imdb_wrapper function.
Finally, our last example of feature selection technique will be a wrapper, an algorithm which relies on scoring a feature subset based on a trained estimator. We could have used forward/backward floating selection algorithms (such as provided by the mlxtend package) but it is actually very long for the time we have in this practical. We will be using one form of backward selection algorithm which runs reasonnably fast, the recursive feature elimination algorithm.
The recursive feature elimination algorithm requires an estimator which can score the individual features and is iterative; at every iteration it fits an estimator, get the scores and drops the step features the less relevant. For the estimator, we will use a LogisticRegression for which a natural scoring of the features is the absolute value (RFE in scikit actually uses the squared value) of the weights. Now, you again have to represent your data with a bag-of-words with {1-2}-grams and a minimal document frequency of 2. Then setup the pipeline for the estimator as a MaxAbsScaler followed by a logistic regression. Then you can create the RFE and I suggest you to use a large step argument otherwise it will take much too long to compute. There is one technical difficulty when using a scikit.Pipeline with RFE which is that RFE expects your classifier to give the feature importances either with a coef_ or feature_importances_ attribute. However, even if you use a LogisticRegression which does have these properties, when put in a Pipeline, you cannot access these attributes by directly asking them to the pipeline object. Fortunately, in the template code, you are given a wrapper class LinearPipeline which solves that problem :
pipeline = Pipeline([
('scaler', MaxAbsScaler()),
('clf', LogisticRegression(solver='liblinear'))
])
wrapped_pipeline = LinearPipeline(classifier, 'clf')
# Will not work
# selector = RFE(pipeline, ...).fit(...)
# Will work
selector = RFE(wrapped_pipeline, ...).fit(...)
At the end, evaluate your training and testing accuracies, estimate the real risk by cross validation and save the selected vocabulary. You can experiment with different feature set size but I warn you this will take much more time. Remember that when performing the cross-validation, you should always include the feature selection part into the pipeline otherwise you would have an optimistic estimate of the risk.
I was able to reach a cross-validated risk of \(90\%\) with a sparsity of \(88\%\) or so.
Variations/Extensions
We already mentionned some variations with respect to the preprocessing (using a stemmer, filtering out stopwords) but we can also vary the vectorization of our documents. Indeed, so far, we simply used bag-of-words which do not carry much about the semantics of the words. For a bag-of-word using bigrams, if you have either “not good”, “not bad”, “not fantastic”, “prettry good”, …, these are just represented with a count in a single dimension like any other bi-grams. There now exists several pre-trained word vectors representations which are known to embed some semantics (see for example fasttext). That could make sense to use these word vectors and to compute, say, the average of the word vectors of a document to get the representation of a sentence (or maybe use other techniques such as paragraph vectors ?).
Visualization of the separation of classes in the feature spaces of a deep neural network
As we saw in the lectures, dimensionality reduction methods can be used to reduce the dimensionality of data down to 2 or 3 dimensions for visual inspection. This is the topic of this section. In particular, Manifold learning techniques (e.g. LLE, t-SNE, U-Map), which focus on preserving local neighborhood relationships, are well suited for this task.
You will be experimenting with the CIFAR-10 dataset that you do not have to download (but it might still be worth consulting the page to get informations about the data). It consists in \(60000\) \(32\times 32\) colored images with \(10\) classes: \(50000\) training images and \(10000\) test images. The 10 classes are airplane, automobile, bird, cat, deer, dog, frog, horse, ship, truck.
You will be given the features extracted from the successive layers of a neural network and you will be applying manifold learning methods to inspect how the classes get separated as we go through the depth of the neural network. You do not need to know much about neural networks, sufficient to say that we expect as we explore a neural network through its depth, we should get features that are more and more class specific. From a very abstract point of view, we can see a deep neural network as a series of modules, performing some mathematical operations on their input and whose output is a representation of the input data. For example, for the mobilenet_v2 network, there are 212 modules taking as input colored images of size \(32 \times 32\).
- What is the dimensionality of the input space ?
Below, we display the representation at some selected module level of the input image of the rooster.
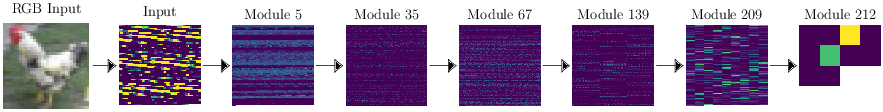
The feature space dimensions are given below :
\[ \begin{array}{cc} \mbox{Module} & \mbox{Feature space size}\\ \mbox{Input} & 3072 \\ 5 & 32768\\ 35 & 147456\\ 67 & 49152 \\ 139 & 36864 \\ 209 & 20480 \\ 212 & 10 \end{array} \]
I used the pretrained mobilenet_v2 network provided by this repository. The features have already been extracted for you with these scripts for only \(5000\) test images since it becomes quite large. For those of you who know about deep learning, the layer 212 is the last linear layer before the softmax and, from the picture above, you see the rooster is classified mainly as a bird (actually with \(92\%\) probability, the green point being for the dog class with probability \(2\%\)).
Now the links to the features :
- cifar10_mobilenet_v2_input.npy a \(5120 \times 3072\) array
- cifar10_mobilenet_v2_labels.npy a \(5120\) array
- cifar10_mobilenet_v2_module_5.npy a \(5120 \times 32.768\) array
- cifar10_mobilenet_v2_module_209.npy a \(5120 \times 20.480\) array
- cifar10_mobilenet_v2_module_212.npy a \(5120 \times 10\) array
Reading these numpy files is as easy as :
import numpy as np
filepath = "/the/path/to/your/npy/file"
data = np.load(open(filepath, 'rb'))
We would like to understand how discriminative these features are with respect to our classes. To do so, we can proceed with dimensionality reduction techniques to get a 2D or 3D representation of our features. I advise you to perform it into two steps: first by using a PCA to reduce the dimensionality (even if you have \(20480\) features, since you have only \(5120\) data, your data does not live in a \(20480\) dimensional space, right ?) and maybe keeping a fraction of \(90\%\) of the variance and then perform the t-SNE down to 2 or 3 dimensions. To perform the PCA and t-SNE, I suggest you to use scikit learn PCA and scikit learn t-SNE
For visualizing the results, you are given the plotter.py script which allows to interactively visualize embeddings. If you have the inputs, labels and embeddings numpy arrays, you simply call :
interactive_show_data(inputs, labels, embeddings)
See the video below for an example using t-SNE on module 209.
As a final note, you could also use this approach as a preliminary step for transfer learning trying to answer the question : are these features still relevant for my dataset which consists in other classes ? You could extract the features for your new dataset, project the data with t-SNE and observe if your new classes get well separated or not with these features. That would give you hints that it’s worth using that network as a base neural network for your dataset.
And as a final final note, if you are interested in visualizing for understanding what a neural network learn, you should have a look to the Exploring Neural networks with activation atlases article.
It is absolutely forbidden to scroll down too much
Solution code
Ahhhhh, I told you ! Anyway, here are possible solution codes.
Dimensionality reduction for learning better predictors
You need the scripts featsel.py, stanford_sentiment.py and utils.py.
Visualization of the separation of classes in the feature spaces of a deep neural network
The code visu.py. To run it, for example (PCA with 10 components keeping \(99.45\%\) of the variance followed by t-SNE)
mylogin@mymachine:~$ python3 scripts/visu.py --data cifar10_mobilenet_v2_module_209.npy --labels cifar10_mobilenet_v2_labels.npy --inputs cifar10_mobilenet_v2_input.npy --num_components 10
To have a plot of the variance you keep with an increasing number of components :
mylogin@mymachine:~$ python3 visu.py --check_variances